Gradient Boosting and XGBoost
Note: This post was originally published on the Canopy Labs website.
XGBoost is an powerful, and lightning fast machine learning library. It’s commonly used to win Kaggle competitions (and a variety of other things). However, it’s an intimidating algorithm to approach, especially because of the number of parameters - and it’s not clear what all of them do.
Although many posts already exist explaining what XGBoost does, many confuse gradient boosting, gradient boosted trees and XGBoost. The purpose of this post is to clarify these concepts. Also, to make XGBoost’s hyperparameters less intimidating, this post explores (in a little more detail than the documentation) exactly what the hyperparameters exposed in the scikit-learn API do.
Contents:
1) Gradient Boosting
2) Gradient Boosted Trees
3) Extreme Gradient Boosting
1. Gradient Boosting
If you are reading this, it is likely you are familiar with stochastic gradient descent (SGD) (if you aren’t, I highly recommend this video by Andrew Ng, and the rest of the course, which can be audited for free). Assuming you are:
Gradient boosting solves a different problem than stochastic gradient descent.
When optimizing a model using SGD, the architecture of the model is fixed. What you are therefore trying to optimize are the parameters, P of the model (in logistic regression, this would be the weights). Mathematically, this would look like this:
\[ F(x \mid P) = \min_{P} Loss(y, F(x \mid P)) \]
Which means I am trying to find the best parameters P for my function F, where ‘best’ means that they lead to the smallest loss possible (the vertical line in \(F(x\mid P)\) just means that once I’ve found the parameters P, I calculate the output of F given x using them).
Gradient boosting doesn’t assume this fixed architecture. In fact, the whole point of gradient boosting is to find the function which best approximates the data. It would be expressed like this:
\[ F(x \mid P) = \min_{F, P} Loss(y, F(x \mid P)) \]
The only thing that has changed is that now, in addition to finding the best parameters P, I also want to find the best function F. This tiny change introduces a lot of complexity to the problem; whereas before, the number of parameters I was optimizing for was fixed (my logistic regression model is defined before I start training it), now, it can change as I go through the optimization process if my function F changes.
Obviously, searching all possible functions and their parameters to find the best one would take far too long, so gradient boosting finds the best function F by taking lots of simple functions, and adding them together.
Where SGD trains a single complex model, gradient boosting trains an ensemble of simple models.
It does this the following way:
Take a very simple model h, and fit it to some data (x, y):
\[ h(x \mid P) = \min_{P} Loss(y, h(x \mid P)) \]
Then, use this trained model to predict an output:
\[ \hat{y} = h(x \mid P) \]
When I’m training my second model, I obviously don’t want it to uncover the same pattern in the data as this first model h; ideally, it would improve on the errors from this first prediction. This is the clever part (and the ‘gradient’ part): this prediction will have some error, \( Loss(y,\hat{y})\). The next model I am going to fit will be on the gradient of the error with respect to the predictions, \( \frac{\partial Loss}{\partial \hat{y}}\).
To think about why this is clever, lets consider mean squared error:
\[ Loss(y, \hat{y}) = MSE(y, \hat{y}) = (y - \hat{y})^2 \]
Calculating this gradient,
\[ \frac{\partial MSE(y, \hat{y})}{\partial \hat{y}} = -2(y - \hat{y}) \propto (y - \hat{y})\]
If for one data point, \(y=1\) and \(\hat{y}=0.6\), then the error in this prediction is \(MSE(1, 0.6) = 0.16\) and the new target for the model will be the gradient, \((y - \hat{y}) = 0.4\). Training a model on this target,
\[ h_{1}(x \mid P) = \min_{P} Loss((y - \hat{y}), h_{1}(x \mid P)) \]
Now, for this same data point, where y=1 (and for the previous model, \(\hat{y} =0.6 \), the model is being trained to on a target of 0.4. Say that it returns \(\hat{y}_{1} = 0.3 \). The last step in gradient boosting is to add these models together. For the two models I’ve trained (and for this specific data point), then
\[ y_{final} = \hat{y} + \hat{y}_{1} = 0.6 + 0.3 = 0.9\]
By training my second model on the gradient of the error with respect to the loss predictions of the first model, I have taught it to correct the mistakes of the first model. This is the core of gradient boosting, and what allows many simple models to compensate for each other’s weaknesses to better fit the data.
I don’t have to stop at 2 models; I can keep doing this over and over again, each time fitting a new model to the gradient of the error of the updated sum of models.
An interesting note here is that at its core, gradient boosting is a method for optimizing the function F, but it doesn’t really care about h (since nothing about the optimization of h is defined). This means that any base model h can be used to construct F.
2. Gradient Boosted Trees
Gradient boosted trees consider the special case where the simple model h is a decision tree. Visually (this diagram is taken from XGBoost’s documentation)):
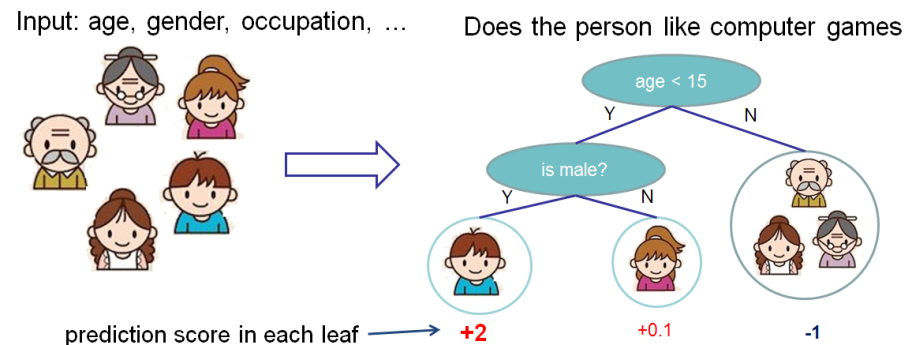
In this case, there are going to be 2 kinds of parameters P: the weights at each leaf, w, and the number of leaves T
in each tree (so that in the above example, T=3 and w=[2, 0.1, -1]).
When building a decision tree, a challenge is to decide how to split a current leaf. For instance, in the above image, how could I add another layer to the (age > 15) leaf? A ‘greedy’ way to do this is to consider every possible split on the remaining features (so, gender and occupation), and calculate the new loss for each split; you could then pick the tree which most reduces your loss.
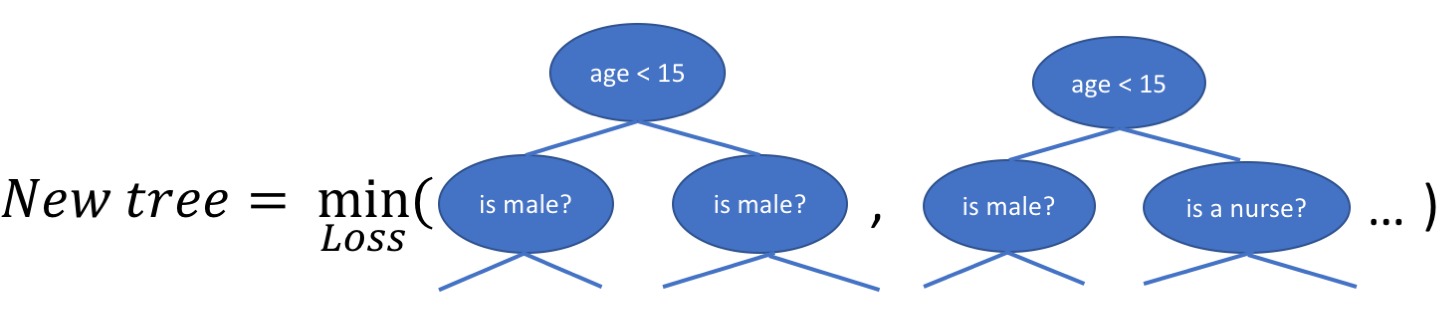
In addition to finding the new tree structures, the weights at each node need to be calculated as well, such that the loss is minimized. Since the tree structure is now fixed, this can be done analytically now by setting the loss function = 0 (see the appendix for a derivation, but you are left with the following):
\[ w_{j} = \frac{\sum_{i \in I_{j}} \frac{\partial loss}{\partial (\hat{y} = 0)}}{\sum_{i \in I_{j}} (\frac{\partial^2 loss}{\partial (\hat{y} = 0)^2}) + \lambda} \]
Where \(I_j\) is a set containing all the instances ((x, y) datapoints) at a leaf, and \(w_j\) is the weight at leaf j. This looks more intimidating than it is; for some intuition, if we consider \(loss=MSE=(y, \hat{y})^2\), then taking the first and second gradients where \(\hat{y} = 0\) yields
\[ w_{j} = \frac{\sum_{i \in I_{j}} y}{\sum_{i \in I_{j}} 2 + \lambda} \]
This makes sense; the weights effectively become the average of the true labels at each leaf (with some regularization from the \(\lambda \) constant).
3. XGBoost (and its hyperparameters)
XGBoost is one of the fastest implementations of gradient boosted trees.
It does this by tackling one of the major inefficiencies of gradient boosted trees: considering the potential loss for all possible splits to create a new branch (especially if you consider the case where there are thousands of features, and therefore thousands of possible splits). XGBoost tackles this inefficiency by looking at the distribution of features across all data points in a leaf and using this information to reduce the search space of possible feature splits.
Although XGBoost implements a few regularization tricks, this speed up is by far the most useful feature of the library, allowing many hyperparameter settings to be investigated quickly. This is helpful because there are many, many hyperparameters to tune. Nearly all of them are designed to limit overfitting (no matter how simple your base models are, if you stick thousands of them together they will overfit).
The list of hyperparameters was super intimidating to me when I started working with XGBoost, so I am going to discuss the 4 parameters I have found most important when training my models so far (I have tried to give a slightly more detailed explanation than the documentation for all the parameters in the appendix).
My motivation for trying to limit the number of hyperparameters is that doing any kind of grid / random search with all of the hyperparameters XGBoost allows you to tune can quickly explode the search space. I’ve found it helpful to start with the 4 below, and then dive into the others only if I still have trouble with overfitting.
3.a. n_estimators (and early stopping)
This is how many subtrees h will be trained. I put this first because introducing early stopping is the most important thing you can do to prevent overfitting. The motivation for this is that at some point, XGBoost will begin memorizing the training data, and its performance on the validation set will worsen. At this point, you want to stop training more trees.
Note that if you use early stopping, XGBoost will return the final model (as opposed to the one with the lowest validation
score), but this is okay since the best model will be this final model minus the additional, overfitting subtrees which
were trained. You can isolate the best model using trained_model.best_ntree_limit in your predict method, as below:
results = best_xgb_model.predict(x_test, ntree_limit=best_xgb_model.best_ntree_limit)
If you are using a parameter searcher like sklearn’s GridSearchCV, you’ll need to define a scoring method which uses the best_ntree_limit:
def best_ntree_score(estimator, X, y):
"""
This scorer uses the best_ntree_limit to return
the best AUC ROC score
"""
try:
y_predict = estimator.predict_proba(X,
ntree_limit=estimator.best_ntree_limit)
except AttributeError:
y_predict = estimator.predict_proba(X)
return roc_auc_score(y, y_predict[:, 1])
3.b. max_depth
The maximum tree depth each individual tree h can grow to. The default value of 3 is a good starting point, and I haven’t found a need to go beyond a max_depth of 5, even with fairly complex data.
3.c. learning rate
Each weight (in all the trees) will be multiplied by this value, so that
\[ w_{j} = \textrm{learning rate} \times \frac{\sum_{i \in I_{j}} \frac{\partial loss}{\partial (\hat{y} = 0)}}{\sum_{i \in I_{j}} (\frac{\partial^2 loss}{\partial (\hat{y} = 0)^2}) + \lambda} \]
I found that decreasing the learning rate very often lead to an improvement in the performance of the model (although it did lead to slower training times).
Because of the additive nature of gradient boosted trees, I found getting stuck in local minima to be a much smaller problem then with neural networks (or other learning algorithms which use stochastic gradient descent).
3.d. reg_alpha and reg_lambda
The loss function is defined as
\[L = \sum_{i=0}^{n} loss(y_{res}, h(x)) + \frac{1}{2}\lambda \sum_{j=1}^{T}w_{j}^2 + \alpha \sum_{j=1}^{T} | w_{j} |\]
reg_alpha and reg_lambda control the L1 and L2 regularization terms, which in this case limit how extreme the
weights at the leaves can become.
These two regularization terms have different effects on the weights; L2 regularization (controlled by the lambda term) encourages the weights to be small, whereas L1 regularization (controlled by the alpha term) encourages sparsity - so it encourages weights to go to 0. This is helpful in models such as logistic regression, where you want some feature selection, but in decision trees we’ve already selected our features, so zeroing their weights isn’t super helpful. For this reason, I found setting a high lambda value and a low (or 0) alpha value to be the most effective when regularizing.
Note that the other parameters are useful, and worth going through if the above terms don’t help with regularization. However, I have found that exploring all the hyperparameters can cause the search space to explode, so this is a good place to start.
Conclusion
Hopefully, this has provided you with a basic understanding of how gradient boosting works, how gradient boosted trees are implemented in XGBoost, and where to start when using XGBoost.
Happy boosting!
Sources
- T. Chen, C. Guestrin, XGBoost: A Scalable Tree Boosting System, 2016
- J. Friedman, Greedy Function Approximation: A Gradient Boosting Machine 1999
- A. Ihler, Ensembles: Gradient Boosting Youtube video, 2012
Appendix
Check out the appendix for more information about other hyperparameters, and a derivation to get the weights.